
The concept of using computers to simulate intelligence and critical thinking was first described by Alan Turing in 1950.

By Dr. Vivek Kaul
Segal-Watson Professor of Medicine
University of Rochester Medical Center

By Sarah Enslin
Physician Assistant
University of Rochester Medical Center

By Dr. Seth A. Gross
Gastroenterology, Advanced Endoscopy
NYU Langone Health
Introduction
Artificial intelligence (AI) was first described in 1950; however, several limitations in early models prevented widespread acceptance and application to medicine. In the early 2000s, many of these limitations were overcome by the advent of deep learning. Now that AI systems are capable of analyzing complex algorithms and self-learning, we enter a new age in medicine where AI can be applied to clinical practice through risk assessment models, improving diagnostic accuracy and workflow efficiency. This article presents a brief historical perspective on the evolution of AI over the last several decades and the introduction and development of AI in medicine in recent years. A brief summary of the major applications of AI in gastroenterology and endoscopy are also presented, which are reviewed in further detail by several other articles in this issue of Gastrointestinal Endoscopy.
The concept of using computers to simulate intelligent behavior and critical thinking was first described by Alan Turing in 1950.1 In the book Computers and Intelligence, Turing described a simple test, which later became known as the “Turing test,” to determine whether computers were capable of human intelligence.2 Six years later, John McCarthy described the term artificial intelligence (AI) as “the science and engineering of making intelligent machines.”3,4

AI began as a simple series of “if, then rules” and has advanced over several decades to include more complex algorithms that perform similarly to the human brain. There are many subfields in AI, akin to specialties in medicine (Table 1), such as machine learning (ML), deep learning (DL), and computer vision.
ML is the use of specific traits to identify patterns that can be used to analyze a particular situation. The machine can then “learn” from and apply that information to future similar scenarios. This prediction tool can be applied dynamically to clinical decision-making to individualize patient care rather than follow a static algorithm.
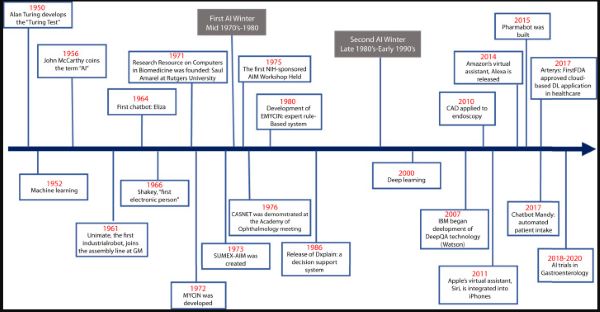
ML has advanced into what is now commonly known as DL, which is composed of algorithms to create an artificial neural network (ANN) that can then learn and make decisions on its own, similar to the human brain.2,5,6 Computer vision is the process by which a computer gains information and understanding from a series of images or videos.7 In this review, we present a brief historical perspective of the advent of AI in medicine (AIM) and its chronological evolution through the last half century leading up to its role in gastroenterology and endoscopy (Fig. 1).
AI in Medicine
Overview
AIM has evolved dramatically over the past 5 decades. Since the advent of ML and DL, applications of AIM have expanded, creating opportunities for personalized medicine rather than algorithm-only–based medicine. Predictive models can be used for diagnosis of diseases, predication of therapeutic response, and potentially preventative medicine in the future.7 AI may improve diagnostic accuracy, improve efficiency in provider workflow and clinical operations, facilitate better disease and therapeutic monitoring, and improve procedure accuracy and overall patient outcomes. The progressive growth and development of the AI platform in medicine is chronicled below and organized by specific time periods of seminal transformation.
The 1950s to 1970s

Early AI was focused on the development of machines that had the ability to make inferences or decisions that previously only a human could make. The first industrial robot arm (Unimate; Unimation, Danbury, Conn, USA) joined the assembly line at General Motors in 1961 and performed automated die casting.8 Unimate was able to follow step-by-step commands. A few years later (1964), Eliza was introduced by Joseph Weizenbaum. Using natural language processing, Eliza was able to communicate using pattern matching and substitution methodology to mimic human conversation (superficial communication),9 serving as the framework for future chatterbots.
In 1966, Shakey, “the first electronic person,” was developed. Created at Stanford Research Institute, this was the first mobile robot to be able to interpret instructions.10 Rather than simply following 1-step commands, Shakey was able to process more complex instructions and carry out the appropriate actions.10 This was an important milestone in robotics and AI.
Despite these innovations in engineering, medicine was slow to adopt AI. This early period, however, was an important time for digitizing data that later served as the foundation for future growth and utilization of AIM. The development of the Medical Literature Analysis and Retrieval System and the web-based search engine PubMed by the National Library of Medicine in the 1960s became an important digital resource for the later acceleration of biomedicine.11 Clinical informatics databases and medical record systems were also first developed during this time and helped establish the foundation for future developments of AIM.11
The 1970s to 2000s

Most of this time period is referred to as the “AI winter,” signifying a period of reduced funding and interest and subsequently fewer significant developments.2 Many acknowledge 2 major winters: the first in the late 1970s, driven by the perceived limitations of AI, and the second in the late 1980s extending to the early 1990s, driven by the excessive cost in developing and maintaining expert digital information databases. Despite the lack of general interest during this time period, collaboration among pioneers in the field of AI continued. This fostered the development of The Research Resource on Computers in Biomedicine by Saul Amarel in 1971 at Rutgers University. The Stanford University Medical Experimental–Artificial Intelligence in Medicine, a time-shared computer system, was created in 1973 and enhanced networking capabilities among clinical and biomedical researchers from several institutions.12 Largely as a result of these collaborations, the first National Institutes of Health–sponsored AIM workshop was held at Rutgers University in 1975.11 These events represent the initial collaborations among the pioneers in AIM.
One of the first prototypes to demonstrate feasibility of applying AI to medicine was the development of a consultation program for glaucoma using the CASNET model.13 The CASNET model is a causal–associational network that consists of 3 separate programs: model-building, consultation, and a database that was built and maintained by the collaborators. This model could apply information about a specific disease to individual patients and provide physicians with advice on patient management.13 It was developed at Rutgers University and was officially demonstrated at the Academy of Ophthalmology meeting in Las Vegas, Nevada in 1976.
A “backward chaining” AI system, MYCIN, was developed in the early 1970s.14 Based on patient information input by physicians and a knowledge base of about 600 rules, MYCIN could provide a list of potential bacterial pathogens and then recommend antibiotic treatment options adjusted appropriately for a patient’s body weight. MYCIN became the framework for the later rule-based system, EMYCIN.11 INTERNIST-1 was later developed using the same framework as EMYCIN and a larger medical knowledge base to assist the primary care physician in diagnosis.11
In 1986, DXplain, a decision support system, was released by the University of Massachusetts. This program uses inputted symptoms to generate a differential diagnosis.3 It also serves as an electronic medical textbook, providing detailed descriptions of diseases and additional references. When first released, DXplain was able to provide information on approximately 500 diseases. Since then, it has expanded to over 2400 diseases.15 By the late 1990s, interest in ML was renewed, particularly in the medical world, which along with the above technological developments set the stage for the modern era of AIM.
From 2000 to 2020: Seminal Advancements in AI

In 2007, IBM created an open-domain question–answering system, named Watson, that competed with human participants and won first place on the television game show Jeopardy! in 2011. In contrast to traditional systems that used either forward reasoning (following rules from data to conclusions), backward reasoning (following rules from conclusions to data), or hand-crafted if-then rules, this technology, called DeepQA, used natural language processing and various searches to analyze data over unstructured content to generate probable answers.16 This system was more readily available for use, easier to maintain, and more cost-effective.
By drawing information from a patient’s electronic medical record and other electronic resources, one could apply DeepQA technology to provide evidence-based medicine responses. As such, it opened new possibilities in evidence-based clinical decision-making.16,17 In 2017, Bakkar et al18 used IBM Watson to successfully identify new RNA-binding proteins that were altered in amyotrophic lateral sclerosis.
Given this momentum, along with improved computer hardware and software programs, digitalized medicine became more readily available, and AIM started to grow rapidly. Natural language processing transformed chatbots from superficial communication (Eliza) to meaningful conversation-based interfaces. This technology was applied to Apple’s virtual assistant, Siri, in 2011 and Amazon’s virtual assistant, Alexa, in 2014. Pharmabot was a chatbot developed in 2015 to assist in medication education for pediatric patients and their parents, and Mandy was created in 2017 as an automated patient intake process for a primary care practice.19,20
DL marked an important advancement in AIM. In contrast to ML, which uses a set number of traits and requires human input, DL can be trained to classify data on its own. Although DL was first studied in the 1950s, its application to medicine was limited by the problem of “overfitting.” Overfitting occurs when ML is too focused on a specific dataset and cannot accurately process new datasets, which can be a result of insufficient computing capacity and lack of training data.21 These limitations were overcome in the 2000s with the availability of larger datasets and significantly improved computing power.
A convolutional neural network (CNN) is a type of DL algorithm applied to image processing that simulates the behavior of interconnected neurons of the human brain. A CNN is made up of several layers that analyze an input image to recognize patterns and create specific filters. The final outcome is produced by the combination of all features by the fully connected layers.5,21 Several CNN algorithms are now available, including Le-NET, AlexNet, VGG, GoogLeNet, and ResNet.22
Applications of DL to Medicine

The application of AI to medical imaging has been suggested to improve accuracy, consistency, and efficiency in reporting. Arterys became the first U.S. Food and Drug Administration–approved clinical cloud-based DL application in health care in 2017. The first Arterys product, CardioAI, was able to analyze cardiac magnetic resonance images in a matter of seconds, providing information such as cardiac ejection fraction. This application has since expanded to include liver and lung imaging, chest and musculoskeletal x-ray images, and noncontrast head CT images.23
DL can be applied to detect lesions, create differential diagnoses, and compose automated medical reports. In 2017, Gargeya and Leng24 used DL to screen for diabetic retinopathy, achieving a 94% sensitivity and 98% specificity with 5-fold cross-validation (area under the curve, .97). Similarly, Esteva et al25 trained a CNN to identify nonmelanoma and melanoma skin cancers with results indicating CNN performance comparable with experts. Weng et al26 showed how a CNN can be used to predict cardiovascular risk in a cohort population. AI was shown to improve accuracy in cardiovascular risk prediction compared with the established algorithm defined by the American College of Cardiology guidelines.26 AI was also applied to reliably predict progression of Alzheimer’s disease by analyzing amyloid imaging data and accurately predicting drug therapy response in this disease.27,28
Applications of AI in Gastroenterology
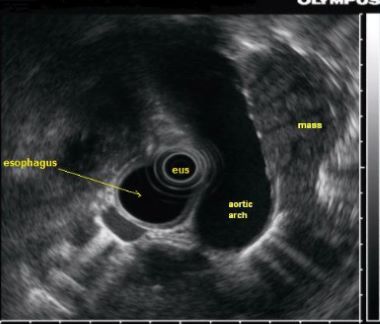
The application of AI in gastroenterology has expanded greatly over the last decade. Computer-assisted diagnosis can be applied to colonoscopy to improve the detection of and differentiation between benign versus malignant colon polyps.7 By using the EUS platform, AI has been used to help differentiate chronic pancreatitis from pancreatic cancer, a common clinical challenge.29,30
DL can also be developed to perform prediction models for prognosis and response to treatment. Several ANNs have been created and tested for diagnosis and prediction models in gastroenterology. In a retrospective study of 150 patients, 45 clinical variables were used to make a diagnosis of GERD with 100% accuracy.31 Rotondano et al32 presented a prospective, multicenter study of 2380 patients in which ANN used 68 clinical variables to predict the mortality in nonvariceal upper GI bleeding with 96.8% accuracy. AI has been used for prognostication of survival in esophageal adenocarcinoma,33 to predict relapse and severity of inflammatory bowel disease,34 and to inform probability of distant metastases in esophageal squamous cell carcinoma, among other similar applications.28 These early studies suggest promise for future application to clinical practice.
AI-Assisted Endoscopy

AI-assisted endoscopy is an evolving field with a promising future. Initial applications included computer-aided diagnosis (CAD) for the detection, differentiation, and characterization of neoplastic and non-neoplastic colon polyps.5,35 A recent randomized controlled trial of 1058 patients demonstrated a significant increase in adenoma detection rates with the use of CAD compared with standard colonoscopy (29% vs 20%, P < .001), with an increased detection of diminutive adenomas (185 vs 102, P < .001) and hyperplastic polyps (114 vs 52, P < .001). There was no statistical difference in the detection of larger adenomas.36 Optical biopsy sampling of colorectal polyps has also been evaluated by several groups using CAD models on images and videos with narrow-band imaging, chromoendoscopy, and endocystoscopy. The diagnostic accuracy of these models has been reported between 84.5% and 98.5%.5 A CNN was developed to determine invasiveness of colorectal mass lesions suspected to be cancer where a diagnostic accuracy of 81.2% was achieved.37
AI has also been applied to improve imaging in Barrett’s esophagus. de Groof et al38 recently developed a CAD system that was 90% sensitive and 88% specific (89% accurate) in classifying images as neoplastic or nondysplastic Barrett’s esophagus. The CAD system had higher accuracy than 53 nonexpert endoscopists (88% vs 73%).38 Similarly, a real-time CAD system was developed and trained using 1480 malignant narrow-band images and 5191 precancerous narrow-band images and was able to differentiate early esophageal squamous cell carcinoma from precancerous lesions with 98% sensitivity and 95% specificity (area under the curve, .989).39
CNN-based models have also been applied to detecting small-bowel capsule endoscopy anomalies and gastric cancer.40,41,42,43,44 Cazacu et al30 reported using ANNs in conjunction with EUS to assist in differentiating chronic pancreatitis from pancreatic cancer, with a sensitivity of 95% and specificity of 94%.
The role of AI in endoscopic practice continues to evolve at a rapid pace and has benefited from the overall technological revolution in the endoscopy and imaging space in recent years. Although much of the technology reported has been proof of concept, 2 systems are approved for use. ENDOANGEL (Wuhan EndoAngel Medical Technology Company, Wuhan, China), a CNN-based system developed in 2019, can provide an objective assessment of bowel preparation every 30 seconds during the withdrawal phase of a colonoscopy, achieving a 91.89% accuracy.45 A recent randomized controlled study used the device to monitor real-time withdrawal speed and colonoscopy withdrawal time and demonstrated significant improvement in adenoma detection rates using ENDOANGEL-assisted colonoscopy versus unassisted colonoscopy (17% vs 8%; odds ratio, 2.18; 95% confidence interval, 1.31-3.62; P = .0026).46
The second system, GI Genius (Medtronic, Minneapolis, Minn, USA), is an AI-enhanced endoscopy aid device developed to identify colorectal polyps by providing a visual marker on a live video feed during endoscopic examination. It is approved for use in Europe and undergoing clinical evaluation in the United States. In a validation study, GI Genius had an overall sensitivity per lesion of 99.7% and detected polyps faster than endoscopists in 82% of cases.47 In a recent randomized controlled trial, Repici et al48 demonstrated a 14% increase in adenoma detection rates using this CAD system.
Conclusion
AI has come a long way from the initial Turing test to its current avatar. As we enter the “roaring twenties,” we are at the dawn of a new era in medicine when AI begins its immersion into daily clinical practice. The potential applications for the role of AI in GI disease currently have no boundaries. These range from improving our diagnostic ability in endoscopy, making endoscopy workflow more efficient, and even helping to more accurately risk stratify patients with common GI conditions such as GI bleeding and neoplasia.
However, AI algorithms and their applications will need further study and validation. Furthermore, additional clinical data will be needed to demonstrate its efficacy, value, and impact on patient care and outcome. Finally, we will need to develop cost-effective AI models and products to allow physicians, practices, and hospitals to incorporate AI into daily clinical use. Physicians should not view this as “human versus machine” but rather a partnership in an effort to further improve clinical outcomes for the patient with GI disease.
See endnotes and bibliography at source.
Originally published by Gastrointestinal Endoscopy 92:4 (October 2020, 807-812) under an Open Access license.


